In the middle of the 20th century, a quiet revolution occurred in the way that scientists understood the building blocks of life. While biologists had long known that all organisms are made up of many microscopic cells, they had relatively little insight into what mechanisms were at work inside the cell wall. For example, while it was known that proteins are a key component in cellular processes, little was known about how cells convert a soup of amino acids into a functional protein. Scientists all over the world began to realize that, in order to understand the nature of the living world, we must first understand the intracellular molecules that compose it.
At first it was sufficient to simply catalog as many of these molecules as possible; however, it soon became apparent that the key is to understand how these molecules interact. It turns out that the thousands of proteins in each cell act together in a giant network that makes up an organism’s operating system. At a basic level, a cell is a computer that uses chemistry instead of electricity, with DNA, RNA, and proteins playing the roles of transistors.
We might then start to think of collections of molecules that share close interactions as forming circuits. These circuits are imbued with particular functions—for example, breaking down sugar or sensing if there are dangerous chemicals nearby. These circuits in turn are connected to other circuits, ultimately forming an architecture that organizes all the processes within a cell. The study of this architecture has led to the development of the field of systems biology, in which biologists work with engineers, physicists, and mathematicians to understand the system-level organization that turns a bag of chemicals into a living cell.
While there is a wide variety of processes currently studied in systems biology, one of the most well-understood is the heat shock response system. We can think of each protein inside of a cell as having some function, and this function depends crucially on how the protein is folded. Sudden changes in temperature can cause proteins to unfold and become dysfunctional, wreaking havoc inside a cell. Without an aggressive response, this will eventually kill the cell. Depending on how large the temperature change is, this can happen in just a few minutes. As most cells have a strong preference for not dying, a sophisticated disaster response system kicks in that rapidly refolds or destroys the unfolded proteins. While every organism so far discovered has a heat shock response system, it is particularly well understood in the bacterium Escherichia coli, better known as E. coli.
Electron microscopy image of E. coli; each bacterium is approximately 2 micrometers long.
National Institute for Allergy and Infectious Diseases
The core player in this process is a protein called a chaperone, whose job it is to find unfolded proteins and refold them. While the chaperone is the star of the show, its sidekick is RpoH, a protein that is in charge of activating other genes—in this case, the genes that make the chaperone. This regulation is essential, as it would be a waste of resources to constantly produce chaperones when the cell doesn’t need them. The question then becomes, how does RpoH know when to make chaperones? Somehow it must sense when the cell is in danger of overheating and respond appropriately.
To do this, RpoH has three different strategies, involving two feedback responses and a feedforward response. Feedback is the process of measuring the output of a system (e.g., unfolded protein), and adjusting the input (e.g., concentration of RpoH) accordingly. E. coli has evolved a clever trick to implement this behavior, by having the RpoH protein bind to chaperones in what we will refer to as sequestration feedback. The key is that the chaperone likes binding to unfolded protein more than it likes binding to RpoH. When the level of unfolded proteins increases, RpoH gets knocked off from the chaperone and is free to initiate the production of more chaperones. When there are more chaperones than unfolded proteins to process, free RpoH will rebind to the excess chaperones and therefore be unable to initiate further production. This process is illustrated graphically below.
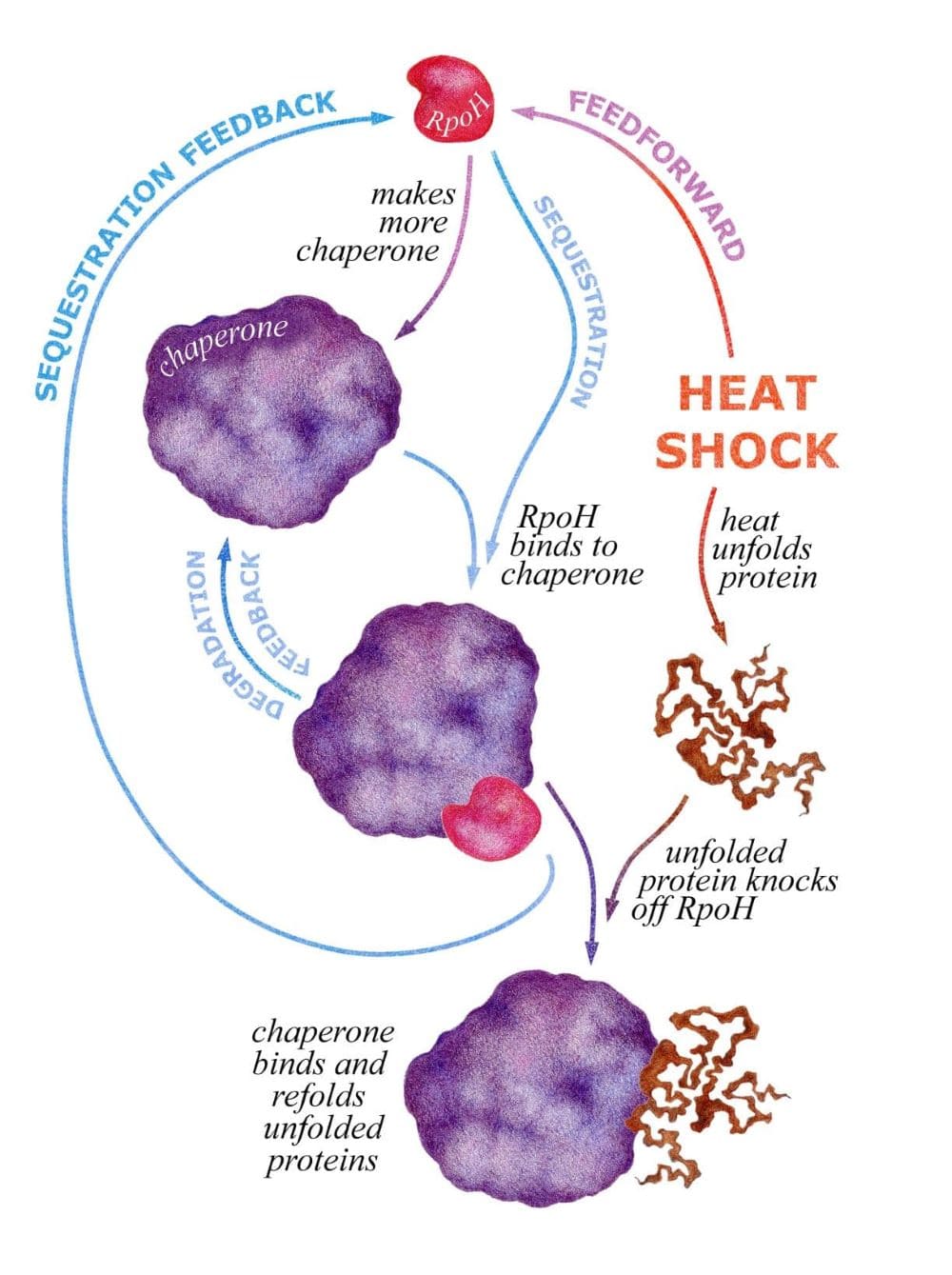
The heat shock response system in E. coli. We see the refolding by chaperones of heat-damaged proteins is regulated by two feedback mechanisms (sequestration and degredation) and a feedforward response. These allow the system to not only respond to heat shock, but do so quickly and efficiently.
While feedback relies on measuring the level of unfolded protein, feedforward acts like an early-warning system. The feedforward mechanism is responsible for directly sensing temperature and increasing the production of new RpoH when things heat up. This allows the system to start responding to heat shock before proteins have even started to unfold, like stocking up on supplies before a big storm. As yet another measure to ensure that the cell is ready to deal with heat shock, the system also has a secondary feedback mechanism that regulates the degradation of RpoH as a function of how much unfolded protein is present, which we refer to as degradation feedback. In normal conditions RpoH is rapidly degraded; however, when heat shock occurs this degradation all but shuts off, leading to a net increase of RpoH in the cell.
In my work, I approach the heat shock system not as a collection of molecular interactions, but as a highly engineered control system. Control systems are present throughout modern life, from cruise control and thermostats to power plants and autonomous vehicles. These systems allow our technology to automatically adapt in unstable conditions and produce consistent behavior. My research applies the mathematical tools developed in engineering to study the regulatory behavior that exists inside cells. Heat shock response is one of the best examples of a biological control system as it serves an essential function inside of each cell, preparing for an unexpected disaster. It is analogous to the fan controller in your computer, without which your computer could overheat and become permanently damaged.
While the primary task of the system is to refold proteins, the cell likely cares about performance metrics like how much energy is used by the circuit, how fast it responds, and how efficient it is. Why would evolution design such a convoluted mechanism to do such a seemingly simple task? To answer this question we must view this system not just as a single circuit, but as one of many possible circuit architectures that might perform the same function. We find from detailed mathematical representations of these architectures that, while some simpler circuits with fewer regulatory steps can perform well on individual performance goals, the natural architecture—with all of its complexity—hits a sweet spot of speed (how fast the system responds), cost (how many proteins the system produces), and efficiency (the fraction of the produced proteins actively contributing to the response, as opposed to sitting idle).
To understand this, it is useful to think of a naive architecture for the system. Imagine that the cell just kept a large number of chaperones around all the time, rather than waiting to make them in response to a change in temperature. This would be the fastest possible system, but it would also be incredibly wasteful. If heat shock is rare, then the cell wastes a lot of energy making proteins that aren’t doing anything useful most of the time. This is why the cell uses feedback to measure the demand for chaperones and try to match up the supply by regulating RpoH. If, however, the cell waits too long to start producing chaperones, it will die. This is where the extra feedback and feedforward mechanisms come into play—they speed up the entire process so the cell can respond to heat shock within just a few minutes of its occurrence.
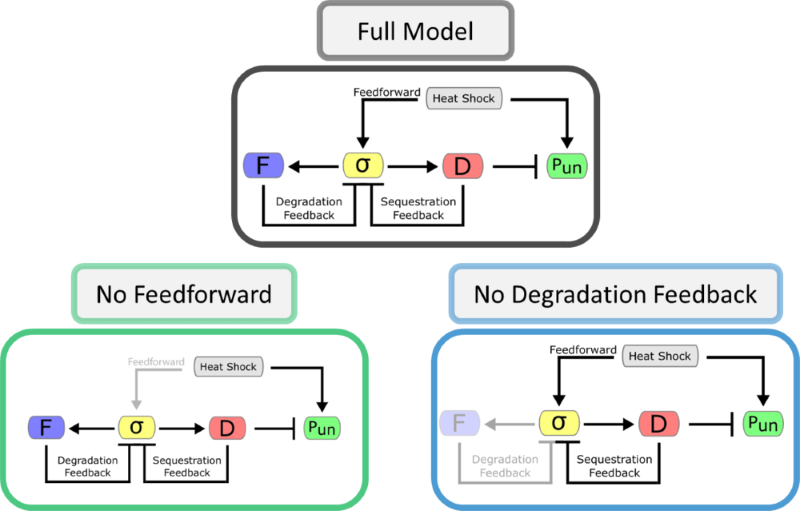
Schematic diagrams of the different system architectures studied in a paper we published in 2018. We compare a full model of the heat shock system to two simpler architectures, one that lacks the feedforward response and another that lacks one of the feedback loops.
Nature is acting like an engineer—rather than optimizing for one particular goal, it has to strike a balance between various performance tradeoffs. To study this quantitatively, I used a mathematical representation of the heat shock system to show that the natural circuit architecture, with its two feedback loops and one feedforward loop, consistently outperforms simpler setups with fewer control mechanisms. For instance, I compared the system’s response time (how long it takes to refold proteins after heat shock) to its efficiency (how many excess chaperones get produced beyond what is needed) and found that simpler architectures can be either faster or more efficiently use resources than the natural one, but generally not both. Because the heat shock response system evolved under strong selective pressure to be fast, robust, and efficient, it is a highly optimized piece of molecular programming. Another example of evolution arriving at a smart solution to a difficult problem!
This is just one example of the variety of functional modules that exist in biology. Life can sometimes feel like the messy result of an aimless process, but perhaps we can develop a coherent theory of design by broadening our vision beyond the details of any one system. Evolution is equal parts hacker and engineer, and unfortunately it does a poor job documenting its work. The goal of systems biologists is to reverse engineer these processes that separate life from non-life. In doing so, we can begin to gain insight into not only what life is made of, but why it is made that way.




