“If understanding everything you needed to know about the brain is a mile, how far have we walked?” asks Jeff Lichtman, Professor of Molecular and Cellular Biology at Harvard University, to his students each year. Some students guess about three-quarters of a mile, others suspect perhaps a half or a quarter of a mile.
Professor Lichtman’s answer? “About three inches.”
Artist's depiction of neuronal activity in the brain
Shutterstock
The incredible complexity of the human brain is what enables us to perform the diverse tasks we do every day, from breathing, walking, and talking to thinking, reading, and problem-solving. At a fundamental level, this complexity is due to the intricate structures of the nearly 100 billion cells that make up our brains: neurons.
One of the many incredible capabilities of our brains is remembering. Take a moment to close your eyes and think about a place you love, somewhere that makes you happy. Perhaps you can remember details of the scenery, some of the sounds, the smells. These memories you just conjured up somehow felt real, but what are they really?
Professor Jeff Lichtman explains the mysteries of the brain
YouTube/National Geographic
There is much scientists have yet to learn about how we remember, but we do have some basic insights. It is widely accepted that the main biological process underlying learning and memory, known as long term potentiation (LTP), involves the strengthening of connections between a specific set of neurons in response to their repeated stimulation. So this memory you just recalled is in fact a collection of cells that have fired together more than others around them, and these cells can therefore send each other stronger signals. But what does it mean for a signal or connection to be stronger? And how do these cells make changes to just one tiny connection out of the nearly hundred trillion in our brain? Before we go into more detail about how this works, we must understand the structure of these neurons that make up our brains.
Most human cells are somewhat round and keep their DNA packed in a compartment near their centers called the nucleus. Neurons, on the other hand, are spindly, irregularly shaped, and highly compartmentalized cells. At their center is the cell body, which contains the nucleus, but stretching out from this cell body are many branch-like projections called dendrites, which receive signals from other neurons, and a single, thin axon through which information is sent to other neurons. Along each dendritic filament are hundreds of tiny protrusions called synapses. These synapses form the junctions where information is transmitted from one neuron to the next via the release and capture of neurotransmitter molecules.
Each of the thousands of synapses on a single neuron is unique. In LTP, during learning and memory formation, specific synapses between repeatedly firing neurons are strengthened. This synaptic strength depends in part on the quantity of neurotransmitters released into the synapse, as well as on the number of receptor molecules that bind these neurotransmitters at the receiving end of the synapse. One can therefore imagine that, in order for a specific synapse to strengthen, extremely localized changes must occur along the length of the dendrite to lead to there being more receptors at the required location.
Membrane receptors and the signaling molecules inside cells with which they communicate all fall into the family of molecules known as proteins. Proteins are created by molecular machines called ribosomes, which translate a messenger RNA (mRNA) into the protein it encodes. In reality, a mRNA is a long, single-stranded molecule containing information from our DNA on how to make a particular protein.
While the transcription of DNA to mRNA occurs in the nucleus, translation of mRNA into protein occurs outside of the nucleus in the cell cytoplasm. Traditionally, mRNA is translated by ribosomes located right outside of the nucleus. In a neuron, however, protein synthesis becomes a bit more puzzling, but, as a result, also more interesting. If mRNA is being made and exported at the nucleus, how do neurons achieve such highly localized changes in protein expression all the way out at individual synapses, hundreds of times farther from the nucleus than where proteins are usually made? Are certain proteins made near the cell body and then shuttled out to where they are needed? Or are some mRNAs and ribosomes transported throughout neurons, such that they can respond rapidly to a local stimulus and make more of the required protein on-site? How do these mRNAs, ribosomes, and/or proteins know exactly where they need to be, and how do they get there?
It is as if someone took a satellite image of the moon and discovered several cars there. With some special cameras and detection techniques, they found that they were American. Yet the question would still remain: were the cars somehow sent from the United States to the moon, or were they made in factories on the moon after instructions for how to build them were delivered? How did the cars or factories and instruction manuals get there?
Scientists have discovered that, indeed, some mRNAs and ribosomes are located out in the dendrites, far from the cell body and nucleus, and they have found evidence that local translation does occur in dendrites and even at or near synapses. However, many questions still remain unanswered. Which specific proteins’ mRNAs are locally translated and in response to what stimuli? What are the time scales of mRNA and ribosome transport, protein translation, and the spread of newly formed proteins? Answering these questions represents a crucial step towards understanding the molecular processes responsible for the functions of our brains.
Scientists have developed several strategies to begin to investigate these questions. Methods such as RNA sequencing reveal information about all of the mRNAs present in a collection of several cells and their relative abundances. Similar methods exist that reveal information about the proteins expressed in a set of cells. In addition to these more global techniques, other methods exist for visualizing and quantifying individual molecules of mRNA, individual proteins, or newly synthesized proteins in single cells. For example, single molecule fluorescence in situ hybridization, or smFISH, enables imaging of individual mRNA molecules in cells using fluorescent tags that act like molecular highlighters to label a specific mRNA of interest. However, knowing the locations of mRNAs or of proteins does not reveal information about where exactly the ribosomes are making the proteins from their mRNA templates.
I work in the Tirrell Lab at Caltech, where recent graduate Dr. Kelly Burke developed a fluorescence assay to detect ribosome interactions with mRNA (FLARIM), which adapts smFISH to visualize and characterize the translation of mRNA. Essentially, to study the translation of a particular protein, we use similar molecular highlighters to tag all of the mRNAs that encode it, as well as the ribosomes interacting with those mRNAs to produce that protein.
More precisely, in FLARIM, fluorescent probes are designed to interact with and label mRNA and a separate set of probes detect the interaction between ribosomes and the mRNA. These probes are all short single strands of DNA that are complementary to, and therefore bind to, sections of the mRNA of interest. Each set of “highlighter” probes fluoresces a unique color and, much like real highlighters, when they label the same molecule, they overlap and give off a new color. Using confocal microscopy, a form of fluorescence microscopy that enables 3D imaging of objects like individual neurons, we can observe bright signal from the mRNAs (red) and see where in the cell it overlaps with with the signal corresponding to the mRNA-bound ribosomes (green) to produce a yellow spot. By counting the number of overlapping yellow spots in our images, we can determine how many of the mRNAs are being translated by ribosomes to make new protein at a given moment in time.
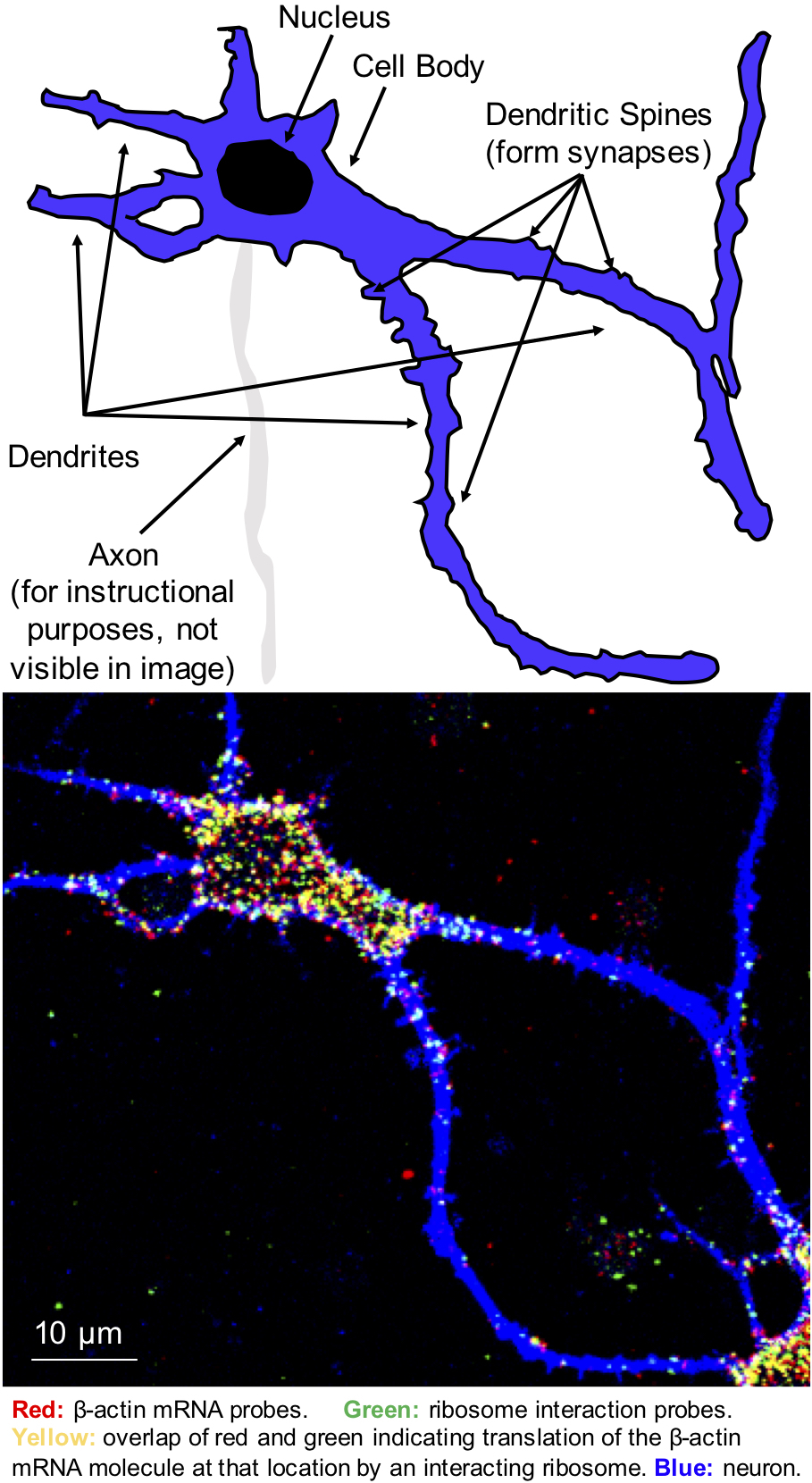
Top: Cartoon depiction of the neuron in the image on the bottom with its various substructures labeled. Bottom: Microscope image showing a neuron in blue. The red dots inside the neurons represent molecules of β-actin mRNA labeled by the mRNA probes. Green dots are produced by the ribosome interaction probes that detect where ribosomes are translating β-actin mRNA. Overlap of red and green dots produces yellow signal, which confirms the interaction of a ribosome with the mRNA molecule of interest at the same location. Red or green dots outside of the cell are background from probes sticking on the surface supporting the neurons.
Sophie Miller, Caltech
The goal of my work is to use FLARIM in neurons to answer outstanding questions about local translation in neurons. I intend to examine translation in dendrites, at synapses, and eventually in axons, which have not been as widely studied. I am currently working with mouse neurons from the hippocampus, a region of the brain involved in learning and memory, to investigate mRNAs that are suspected to be locally synthesized in dendrites. A couple of these genes, such as Arc and CamKIIα, are of particular interest. Arc interacts with other proteins in neurons to help regulate the levels of certain receptors at synapses. CamKIIα, on the other hand, chemically tags receptors and other proteins to make some receptors bind neurotransmitters more readily or to increase the number of receptors at synapses. By causing changes in the amount or type of receptors at synapses, these genes help alter synaptic strength and are therefore implicated in LTP. Intriguingly, both Arc and CamKIIα have both been shown to be necessary for proper learning and special memory formation, and mRNAs for each have been found in dendrites.
Thus far, we have shown that FLARIM works in neurons, and we have used it to detect translation of a widely expressed structural protein in cells, β-actin, as well as of CamKIIα and Arc in the cell body and in dendrites. Furthermore, we have been able to confirm a known increase in translation of CamKIIα and Arc that occurs in response to neuronal activation by a growth factor protein known as brain-derived neurotrophic factor (BDNF).
With these promising initial results for the application of FLARIM to neurons, we are now looking towards investigating proteins whose patterns of translation are less well understood. In particular, we are interested in using FLARIM to determine the extent of local translation of glutamate receptors. Due to their prevalence and role in binding glutamate, the most abundant neurotransmitter in the brain, these receptors are implicated not only in learning and memory formation, but also in various neurological conditions, including epilepsy, Parkinson’s disease, and multiple sclerosis. We will use different methods of stimulating the cells to see what may lead to an increase or decrease in the local translation of these glutamate receptor subunits.
Today, many diseases, including many cancers and brain diseases, are treated with therapies that have been found to cure patients but whose mechanisms of action are unknown. This often leads to undesirable, and at times even harmful or deadly, side effects. By gaining a deeper understanding of how the molecules and cells in our brain actually work and interact with one another, we can intelligently develop new, more effective cures to neurological diseases and psychological disorders.
One of the most exciting and remarkable things about this field of research is that, fundamentally, my brain is learning about itself through my work. As you reach the end of this article, some of the neurons in your brain have undergone the processes I have described here. Perhaps they have made new connections, or strengthened existing ones, and formed new memories by encoding information about themselves in your brain.




